Hello,
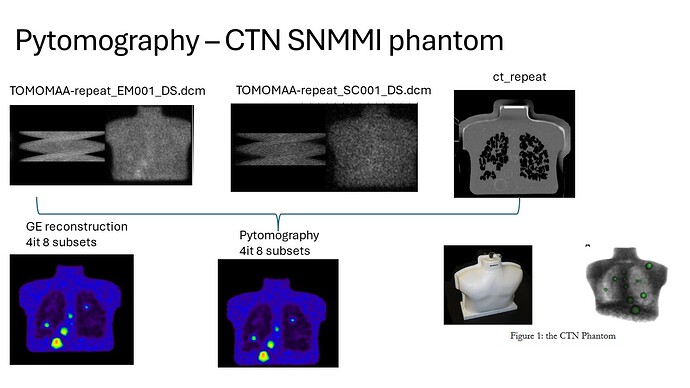
First, thanks for sharing this code/program; I think it is very useful. I managed to get the Dicom test data running. I am trying to run GE data, acquired on an NM/CT 860 with the LEHRS collimators, of a CTN anthropomorphic chest phantom filled with Tc99m and a ratio @4/1 lesion to the background.
I have CT files, ATT map and two files with the projection data for the EM and SC windows. We would like to explore different reconstructions offered by pytomography and use clinical data as well (not only with Tc99m but also with Lu177 and other isotopes).
My questions are:
Do you have any examples of how to reconstruct GE scanner data with the two separate files? What collimator shall I use from the library (G8-LHRS?)? Can you do reconstructions without PSF modelling? And last… in the dicom example, the Sy-ME collimator used returns:
kernel_dimensions = 2D
min_sigmas = 3
sigma_fit_params = [0.03161992134409504, 0.1248503046423388, 0.0]
What do the parameters mean? We would like to explore in the future measured collimator PSF at different distances and implement it with pytomography. Would that be possible?
Thanks a lot for your help
(If you want me to upload our data let me know)